
Image source: Design Cells/iStock/Getty Images
Benchmarking human whole genome and RNA sequencing using Nvidia Ampere A100 GPUs
Nvidia Ampere A100 GPUs exploit wide SIMT architecture to accelerate world’s most demanding HPC and AI workloads, and one of the key application in HPC is sequencing of human genome and transcriptome. This blog aims to measure runtime required for genome and transcriptome sequencing using GPUs to help understand how quickly sequence analysis can be done.
PARABRICKS DNA FQ2BAM pipeline
Dataset:
- Reference genome: GRCh38
- Whole-genome sequencing run: ERR194147 (Illumina HiSeq 2000 paired end sequencing, ~50x coverage)
Results:
Table
Execution Time
| Number of GPUs | Execution Time(s) (alignment-phase BWA-mem) | Execution Time(s) (Overall) |
|---|---|---|
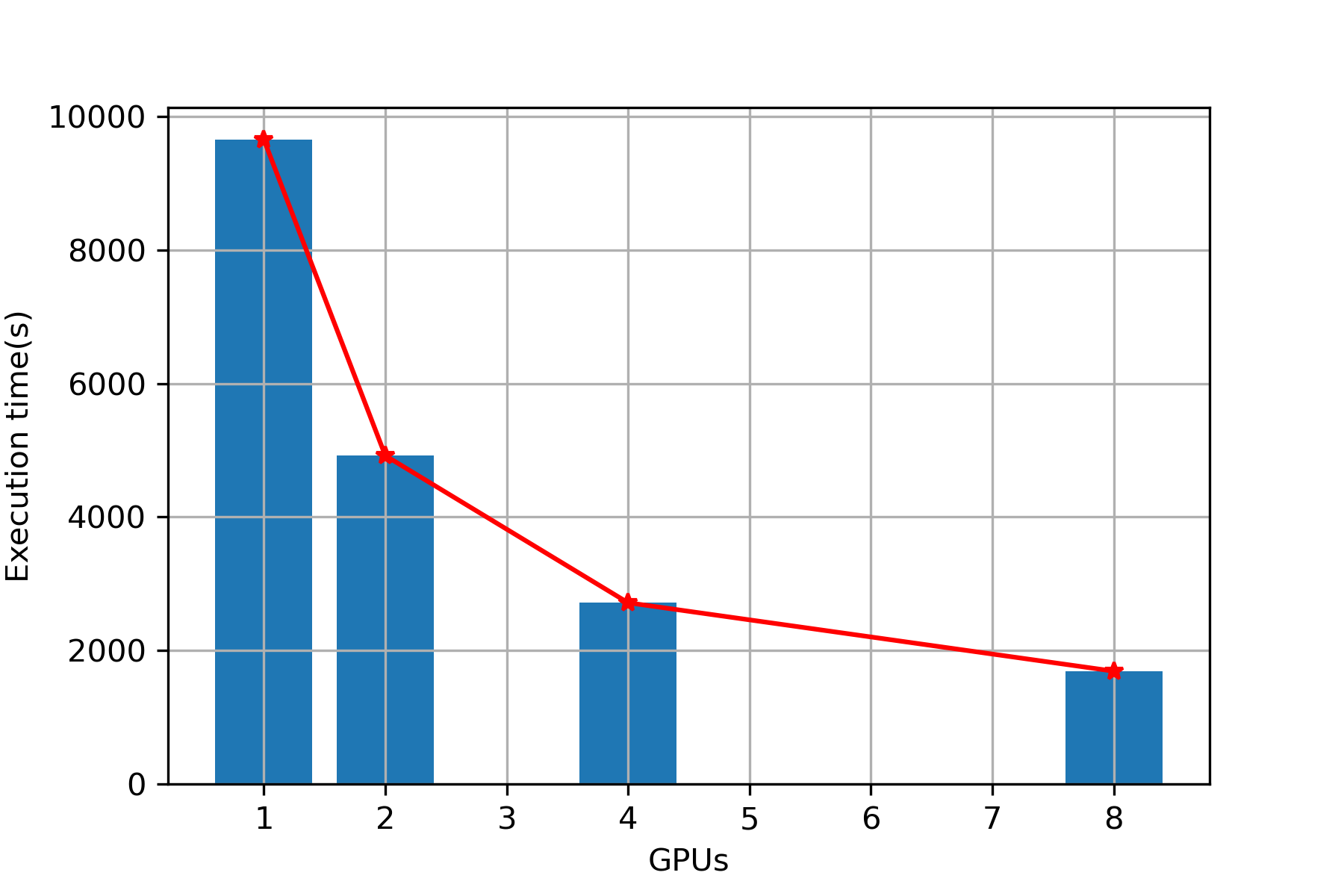
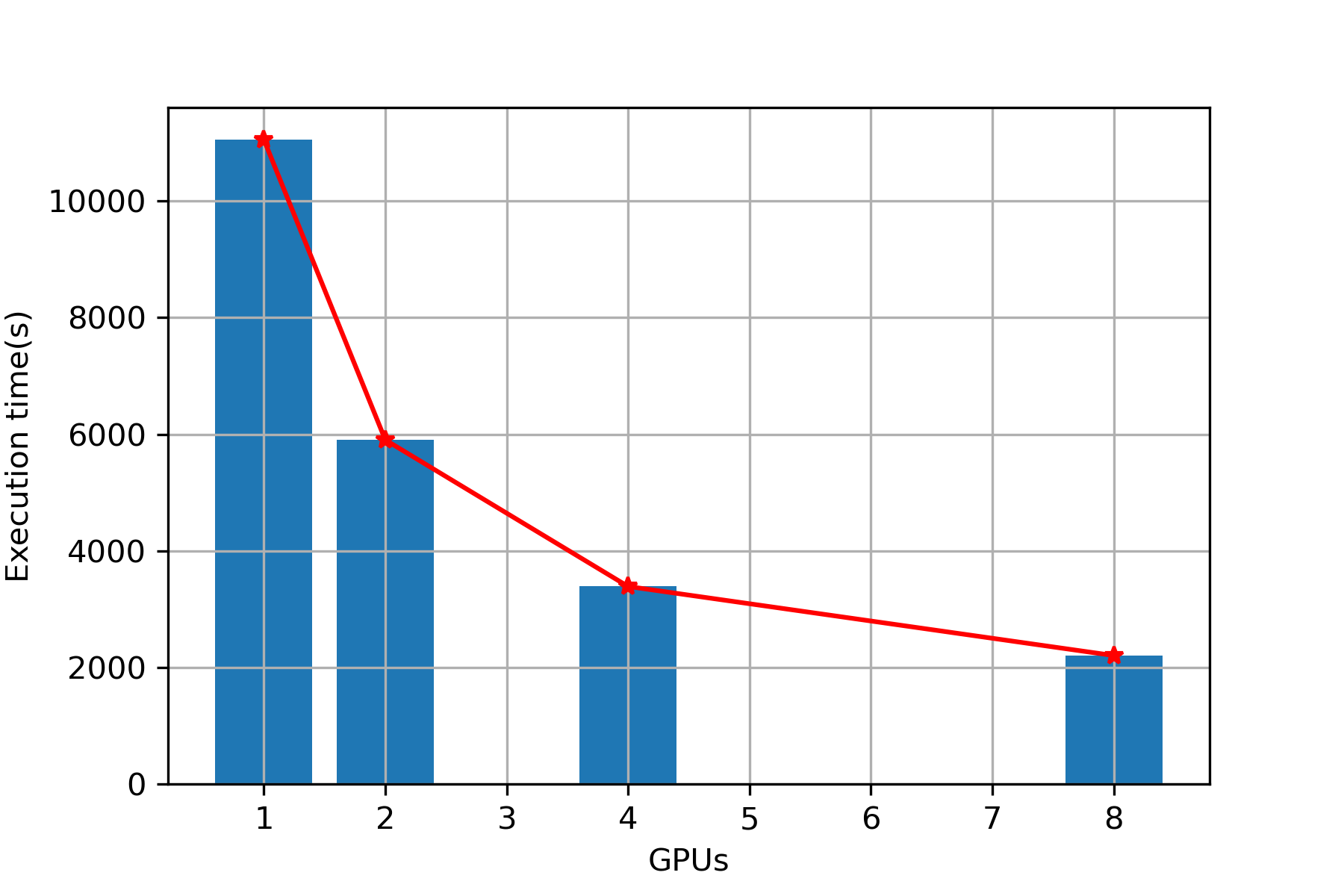
| 1 | 9657 | 11053 |
| 2 | 4924 | 5903 |
| 4 | 2720 | 3389 |
| 8 | 1695 | 2204 |
Plot (Sequence alignment)
Execution Time
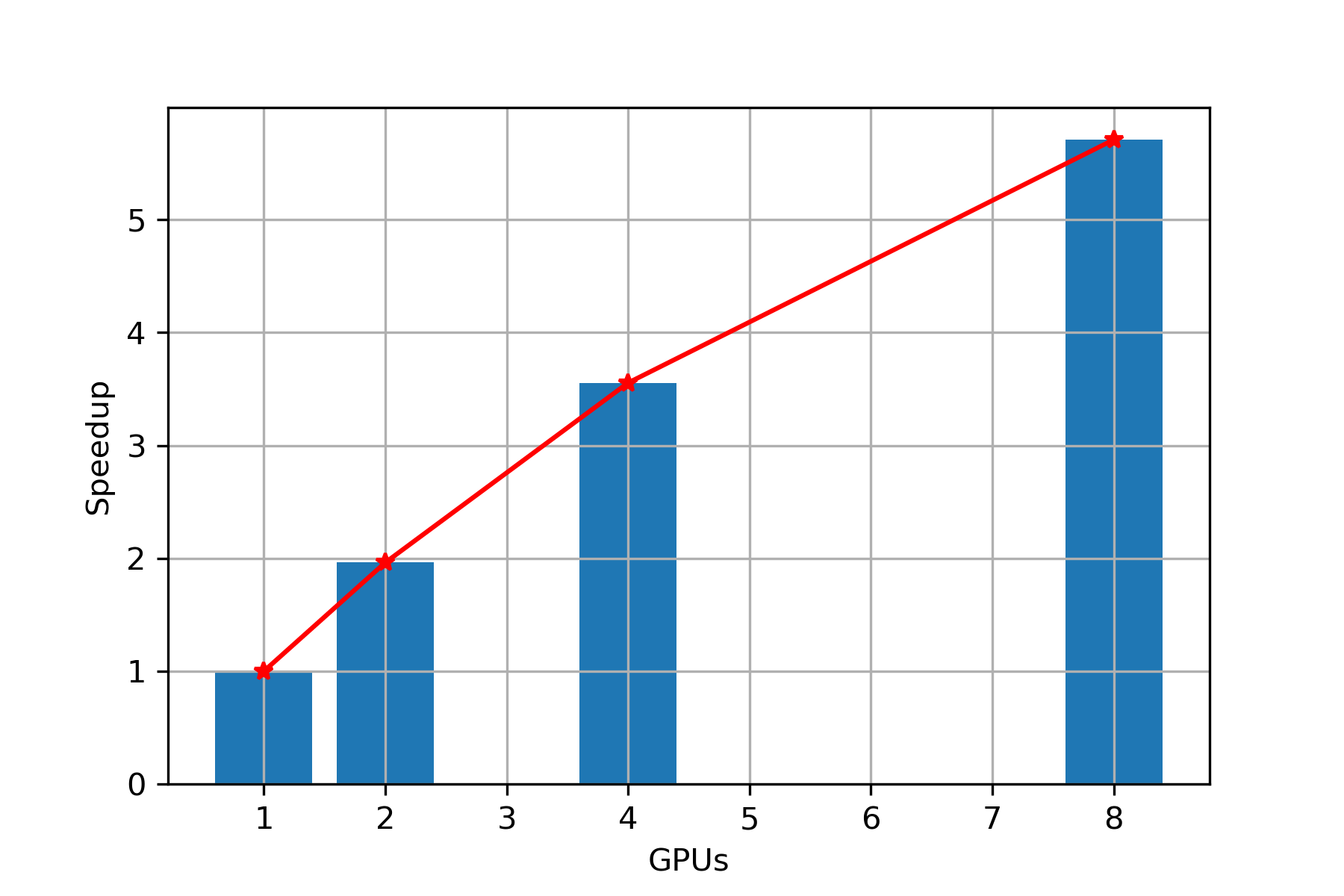
Speed Up
Plot (Overall)
Execution Time
Speed Up
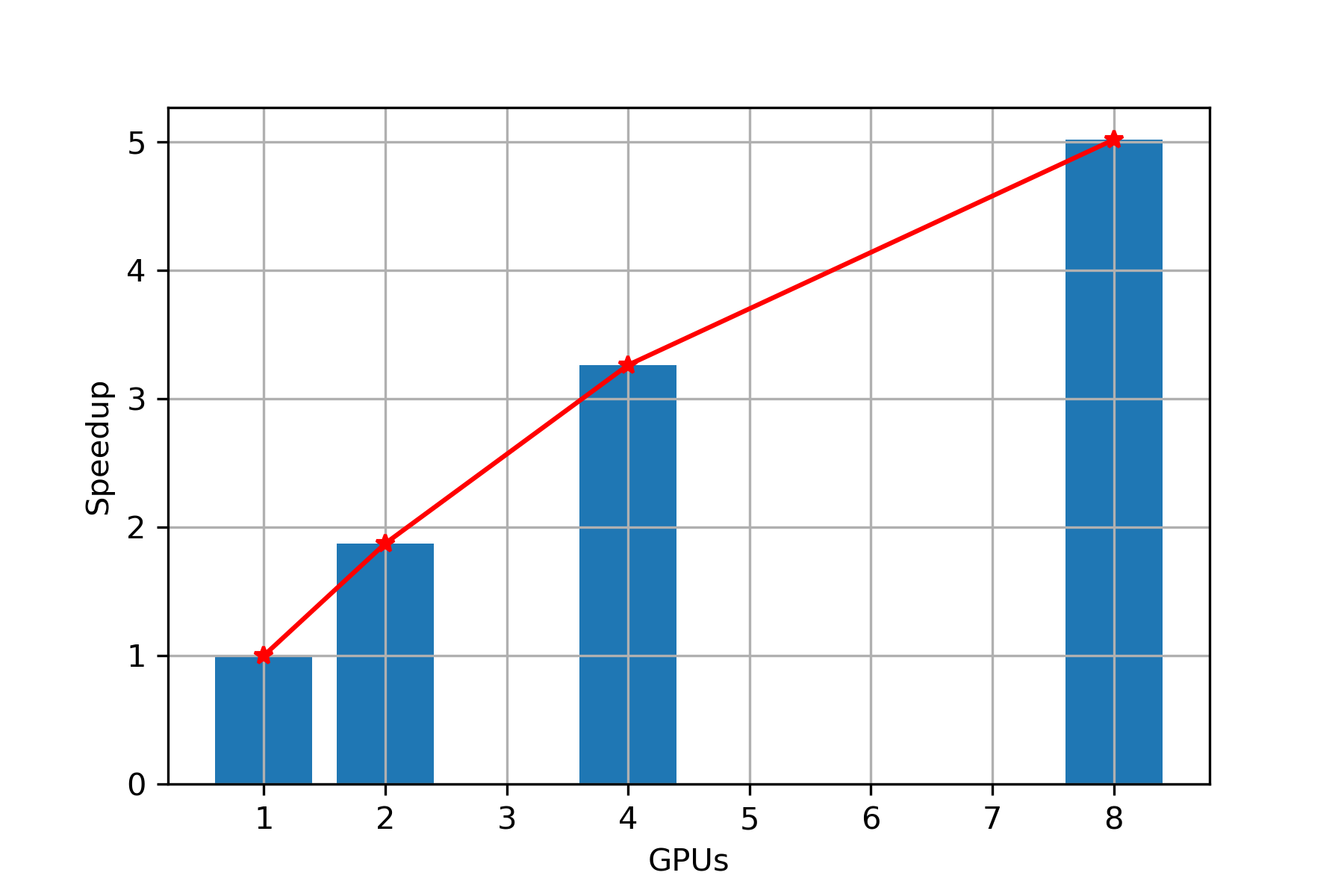
Scaling: From Figure 4, we can clearly observe that with increase in number of GPUs, we are getting almost linear speedup, hence we can conclude that PARABRICKS DNA pipeline is strong scalable upto 8 GPUs.
PARABRICKS RNA FQ2BAM pipeline
Dataset:
Results
Table
Execution Time
| Number of GPUs | Execution Time(s) (alignment-phase STAR) | Execution Time(s) (Overall) |
|---|---|---|
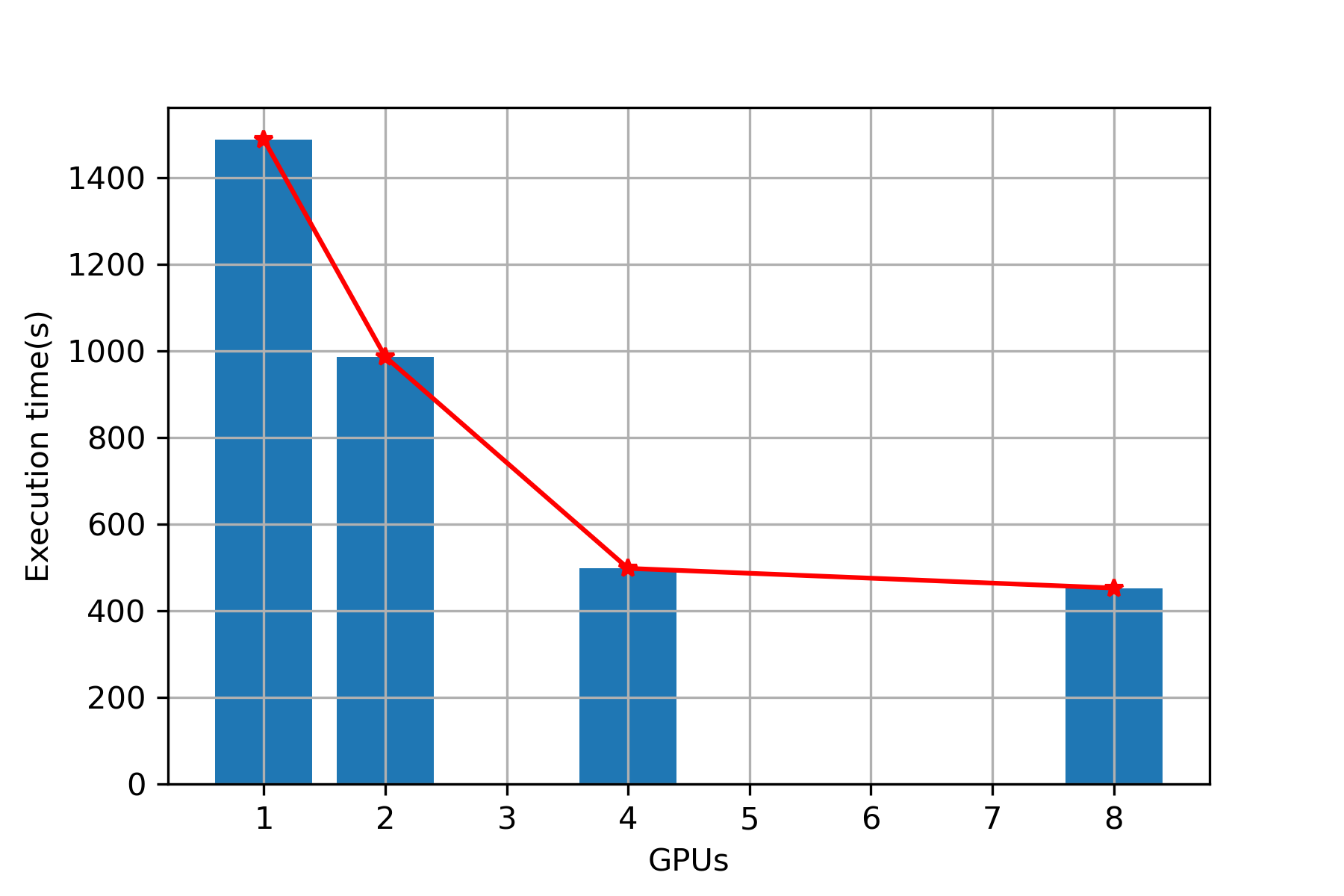
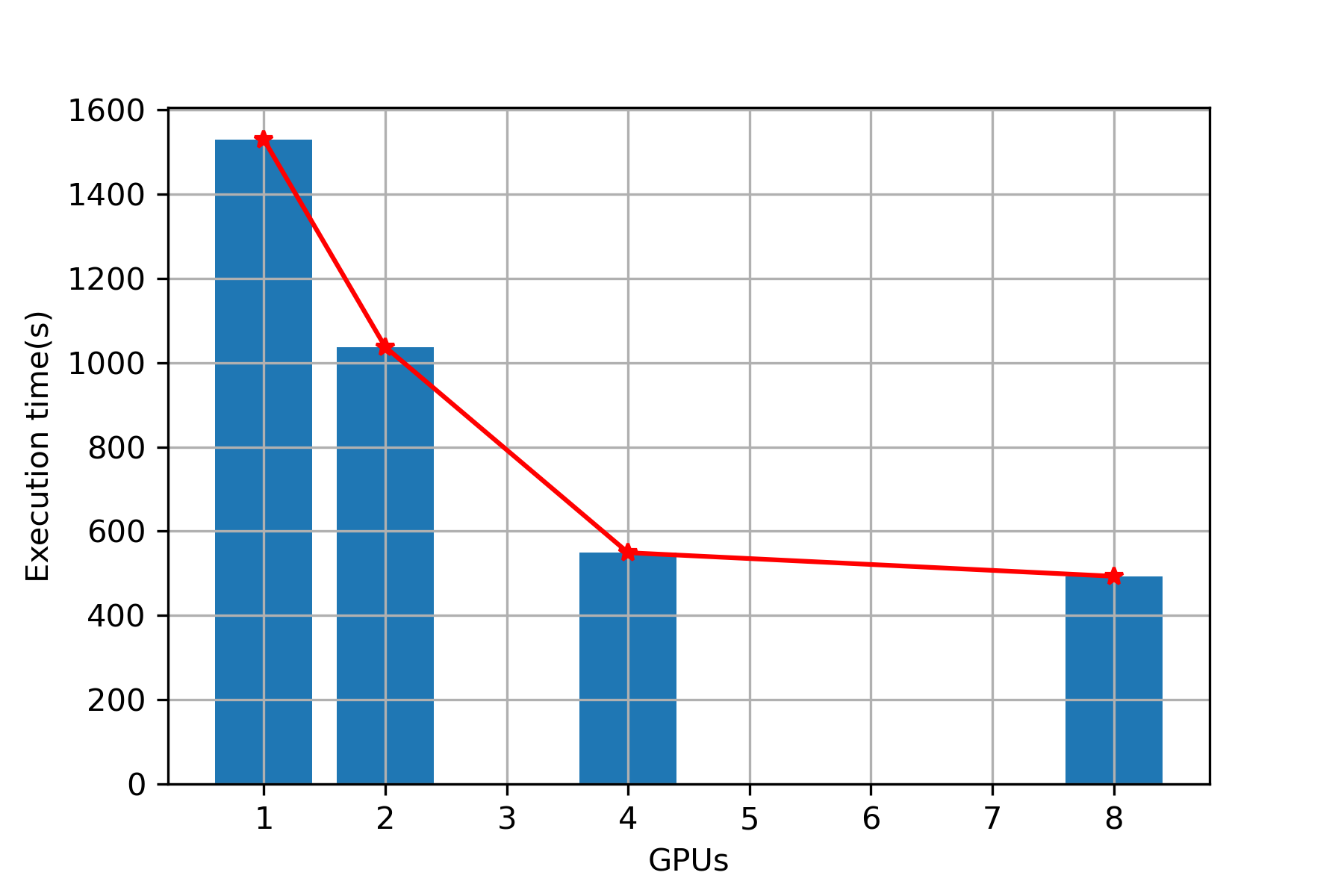
| 1 | 1488 | 1529 |
| 2 | 987 | 1037 |
| 4 | 498 | 549 |
| 8 | 453 | 493 |
Plot (Sequence alignment)
Execution Time
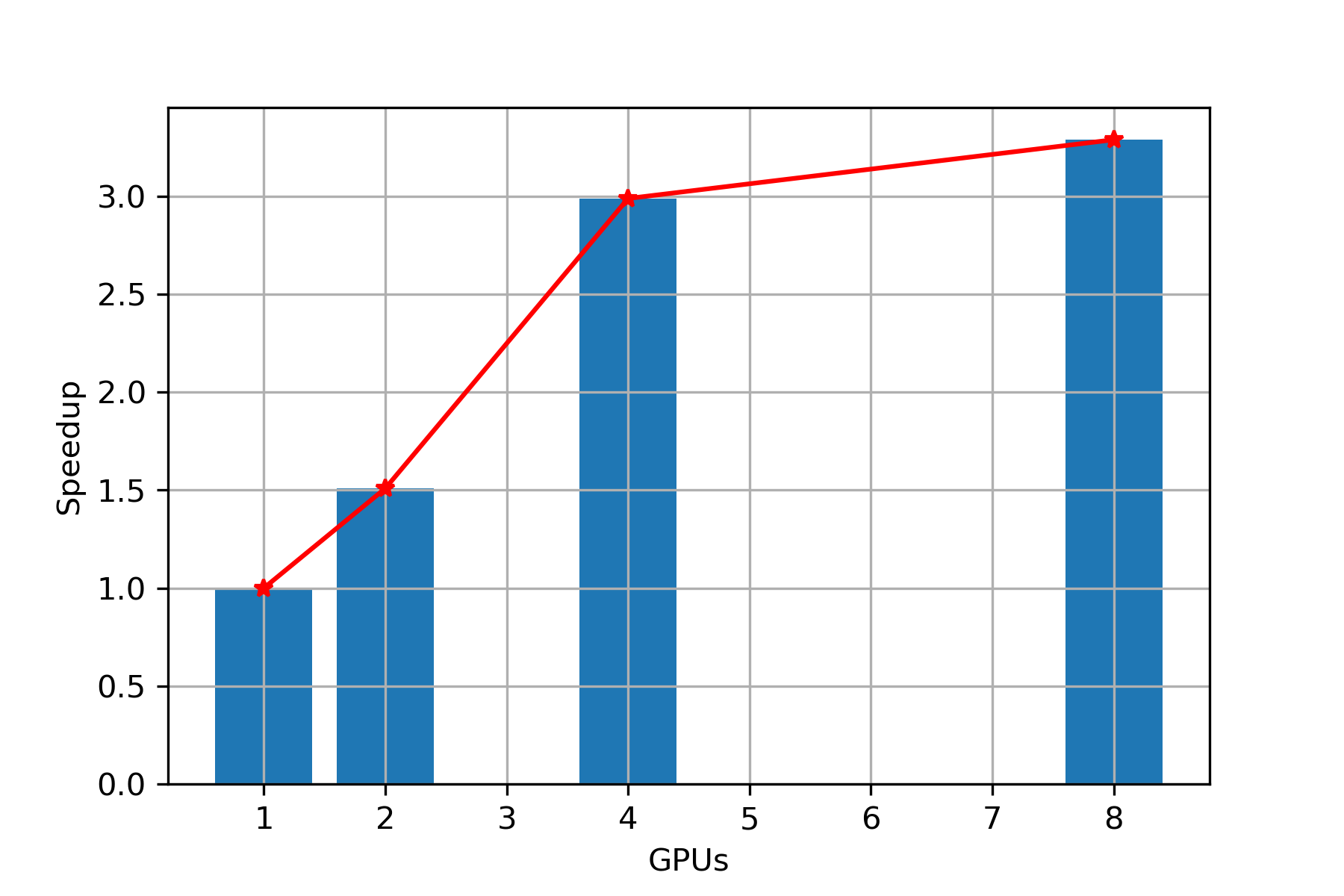
Speed Up
Plot (Overall)
Execution Time
Speed Up
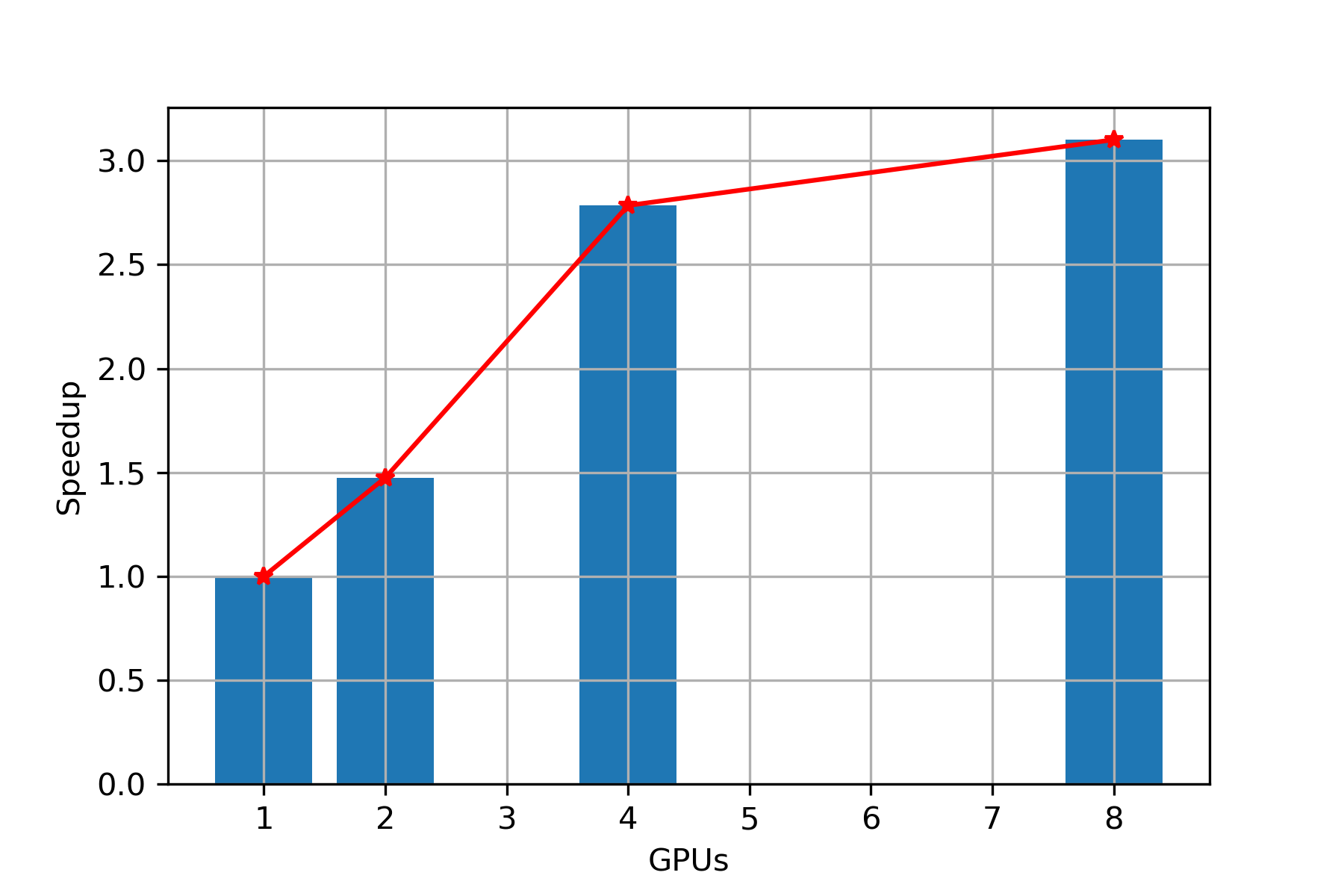
Scaling: In Figure 8, we observe that while keeping data set fixed and with increase in number of GPUs, we are getting almost linear scaling up to 4 GPUs and then the speedup growth reduces. Hence, PARABRICKS RNA pipeline is showing good strong scaling up to 4 GPUs.
Experimental Configuration
- Compute Cluster: PARAM-SIDDHI AI
- Compute Node: Single compute node (Nvidia DGX A100 320GB)
GPU: 8X A100-SXM4-40GB
CPU: Dual AMD EPYC 7742 64-Core Processor
RAM: 1 TB
Storage: 8 PB (lustre fs) - PARABRICKS: Nvidia Clara Parabricks: 3.6.0-1
References
[1]. Nvidia Clara Parabricks 3.6.0-1 [Documentation]
[2]. Param Siddhi supercomputer [DST]
[3]. Nvidia Ampere A100 GPU [Nvidia]
